PD, Dose-Response - Count
Overview
This document contains exploratory plots for count PD data as well as the R code that generates these graphs. The plots presented here are based on simulated data (see: PKPD Datasets). Data specifications can be accessed on Datasets and Rmarkdown template to generate this page can be found on Rmarkdown-Template. You may also download the Multiple Ascending Dose PK/PD dataset for your reference (download dataset).
Setup
# remove reference to home directory in libPaths
.libPaths(grep("home", .libPaths(), value=TRUE, invert=TRUE))
.libPaths(grep("usr", .libPaths(), value=TRUE, invert=TRUE))
# add localLib to libPaths for locally installed packages
.libPaths(c("localLib", .libPaths()))
# will load from first filepath first, then look in .libPaths for more packages not in first path
# version matches package in first filepath, in the case of multiple instances of a package
# library(rmarkdown)
library(gridExtra)
library(grid)
library(ggplot2)
library(dplyr)
library(RxODE)
library(caTools)
#flag for labeling figures as draft
draft.flag = TRUE
## ggplot settings
theme_set(theme_bw(base_size=12))Define Useful Graphics Functions
# annotation of plots with status of code
AnnotateStatus <- function(draft.flag, log.y=FALSE, log.x=FALSE, fontsize=7, color="grey") {
x.pos <- -Inf
if (log.x)
x.pos <- 0
y.pos <- -Inf
if (log.y)
y.pos <- 0
if(draft.flag) {
annotateStatus <- annotate("text",
label="DRAFT",
x=x.pos, y=y.pos,
hjust=-0.1, vjust=-1.0,
cex=fontsize,
col=color, alpha=0.7, fontface="bold")
} else {
annotateStatus <- NULL
}
return(annotateStatus)
}Load Dataset
my.data <- read.csv("../Data/Multiple_Ascending_Dose_Dataset2.csv")
my.data$TRTACT <- factor(my.data$TRTACT,levels = c("Placebo",paste(unique(my.data$DOSE[my.data$DOSE!=0]),"mg")))
my.data$DAY <- floor(my.data$NOMTIME/24)
my.data$DAY_label <- paste("Day",floor(my.data$DAY))
my.data$DAY_label[my.data$DAY<0]<-"Baseline"
my.data$DAY_label <- factor(my.data$DAY_label,
levels = c("Baseline",paste("Day",sort(unique(floor(my.data$DAY))))))
my.data$DAY[my.data$DAY>5] <- 5Provide an overview of the data
Summarize the data in a way that is easy to visualize the general trend of PD over time and between doses. Using summary statistics can be helpful, e.g. Mean +/- SE, or median, 5th & 95th percentiles.
Count over time, colored by Dose, median, 5th & 95th percentiles by nominal time
data_to_plot <- my.data[my.data$CMT==4,]
data_to_plot$TRTACT <- factor(data_to_plot$TRTACT, levels = rev(levels(data_to_plot$TRTACT)))
gg <- ggplot(data = data_to_plot,
aes(x=NOMTIME/24,y=LIDV, color = TRTACT, fill = TRTACT))+theme_bw()
gg <- gg + stat_summary(geom="errorbar",
fun.data=function(y){
data.frame(y=median(y),
ymin=quantile(y,0.025),
ymax=quantile(y,0.975))
}, size = 1, width = 0.4, position = position_dodge(-0.4), alpha = 0.5)
gg <- gg + stat_summary(geom="point", fun.y=median, size = 2, position = position_dodge(-0.4), alpha = 0.5)
# gg <- gg + geom_boxplot(aes(group = interaction(time,TRTACT)), width = 0.2, position = position_dodge(-0.4), fill = NA)
gg <- gg + stat_summary(aes(group = TRTACT), geom="line",fun.y=median, size = 1, position = position_dodge(-0.4))
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Time (days)") + scale_x_continuous(breaks=seq(-1,8,1))
gg <- gg + ylab("Count")
# gg <- gg + geom_smooth( method = "glm",method.args=list(family=poisson), position = position_dodge(-0.4))
gg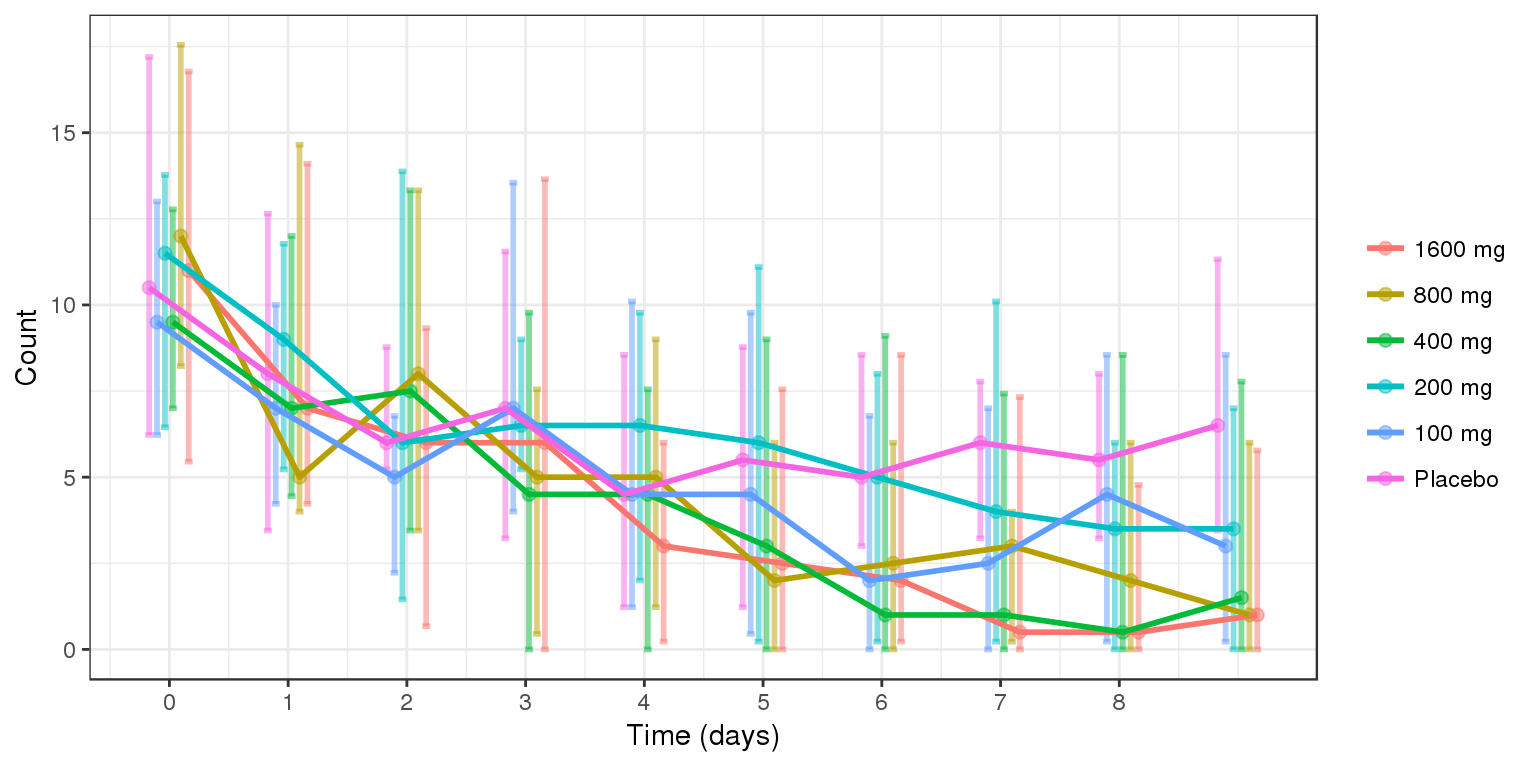
Count over time, faceted by Dose, median, 5th & 95th percentiles by nominal time
data_to_plot <- my.data[my.data$CMT==4,]
gg <- ggplot(data = data_to_plot,
aes(x=NOMTIME/24,y=LIDV))+theme_bw()
gg <- gg + stat_summary(geom="errorbar",
fun.data=function(y){
data.frame(y=median(y),
ymin=quantile(y,0.025),
ymax=quantile(y,0.975))
}, size = 1)
gg <- gg + stat_summary(geom="point", fun.y=median, size=2)
gg <- gg + stat_summary(geom="line", fun.y=median)
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Time (days)") + scale_x_continuous(breaks=seq(-1,8,1))
gg <- gg + facet_grid(~TRTACT)
gg <- gg + ylab("Count")
gg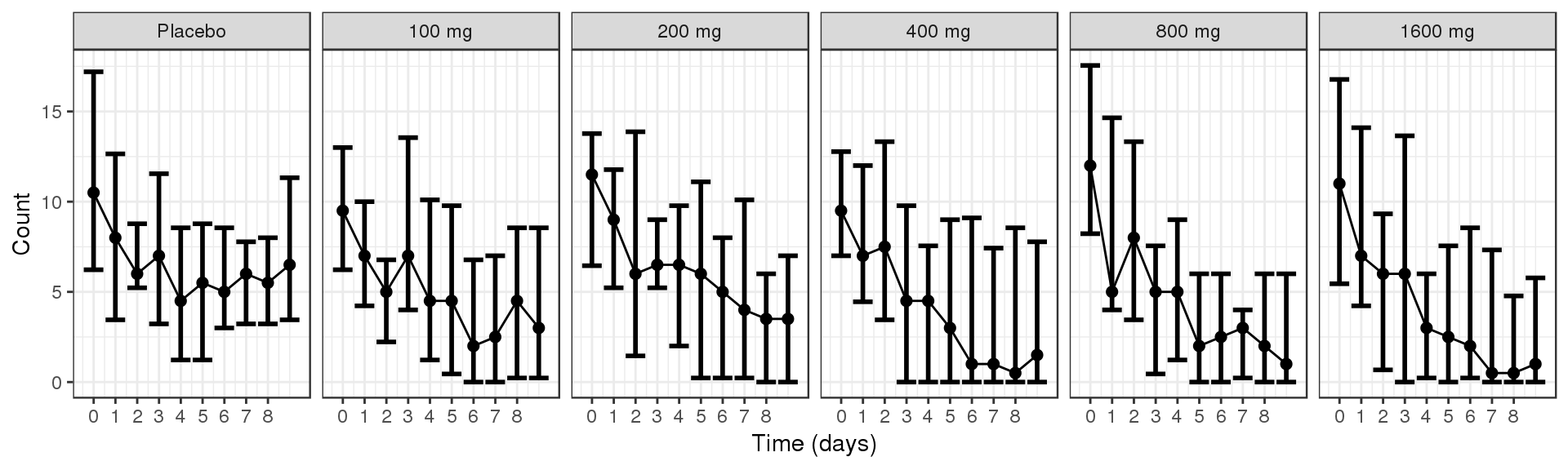
Percent of subjects by Count over time, faceted by dose
data_to_plot <- my.data[my.data$CMT==4,]
# data_to_plot$DAY_label <- factor(data_to_plot$DAY_label, levels = rev(sort(unique(data_to_plot$DAY_label))))
# data_to_plot$TRTACT<- factor(data_to_plot$TRTACT, levels = rev(levels(data_to_plot$TRTACT)))
data_to_plot$Count <- factor(data_to_plot$LIDV, levels = rev(sort(unique(data_to_plot$LIDV))))
gg <- ggplot(data = data_to_plot, aes(x = factor(CYCLE), fill = Count)) + theme_bw()
gg <- gg + geom_bar(position = "fill") + scale_y_continuous(labels = scales::percent)
gg <- gg + ylab("Percent of Subjects") + xlab("Time (days)") + guides(fill=guide_legend(""))
gg + facet_grid(.~TRTACT) + scale_fill_grey(start = 0.8, end = 0.2, breaks = rev(seq(0,18,6)))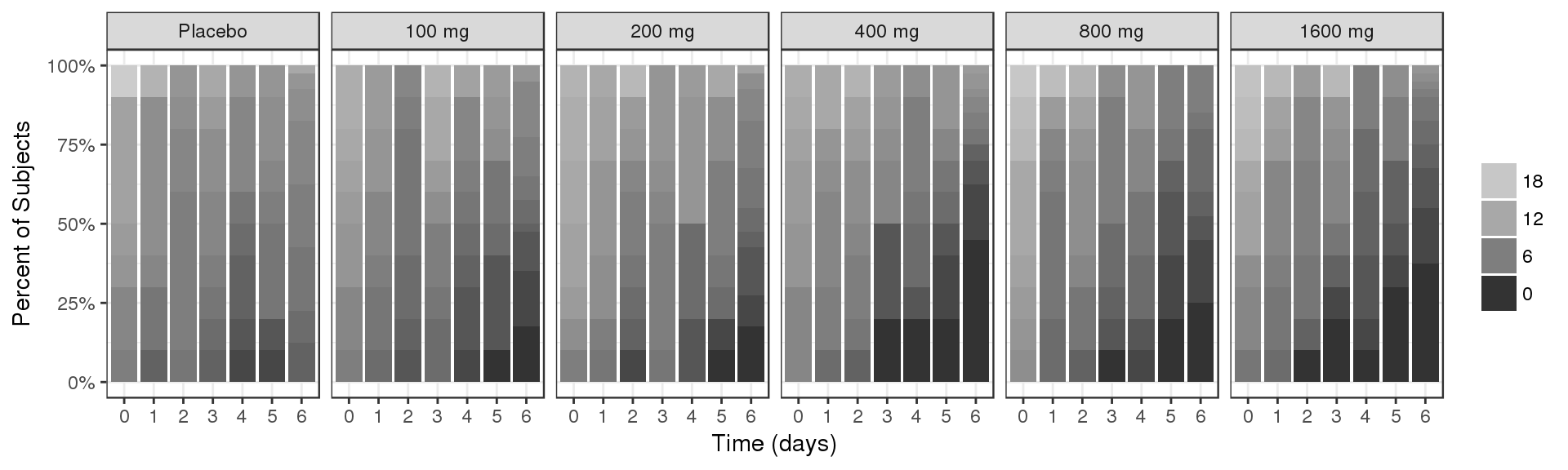
Explore variability
Use spaghetti plots to visualize the extent of variability between individuals. The wider the spread of the profiles, the higher the between subject variability. Distinguish different doses by color, or separate into different panels. If coloring by dose, do the individuals in the different dose groups overlap across doses? Does there seem to be more variability at higher or lower concentrations?
Spaghetti plots of Count over time, faceted by dose
data_to_plot <- my.data[my.data$CMT==4,]
gg <- ggplot(data = data_to_plot,
aes(x=CYCLE,y=LIDV))+theme_bw()
gg <- gg + geom_point( size=2, alpha = 0.5) + geom_line( aes(group = ID), alpha = 0.5)
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Time (days)")+ scale_x_continuous(breaks = seq(-1,8,1))
gg <- gg + facet_grid(~TRTACT)
gg <- gg + ylab("Count")
gg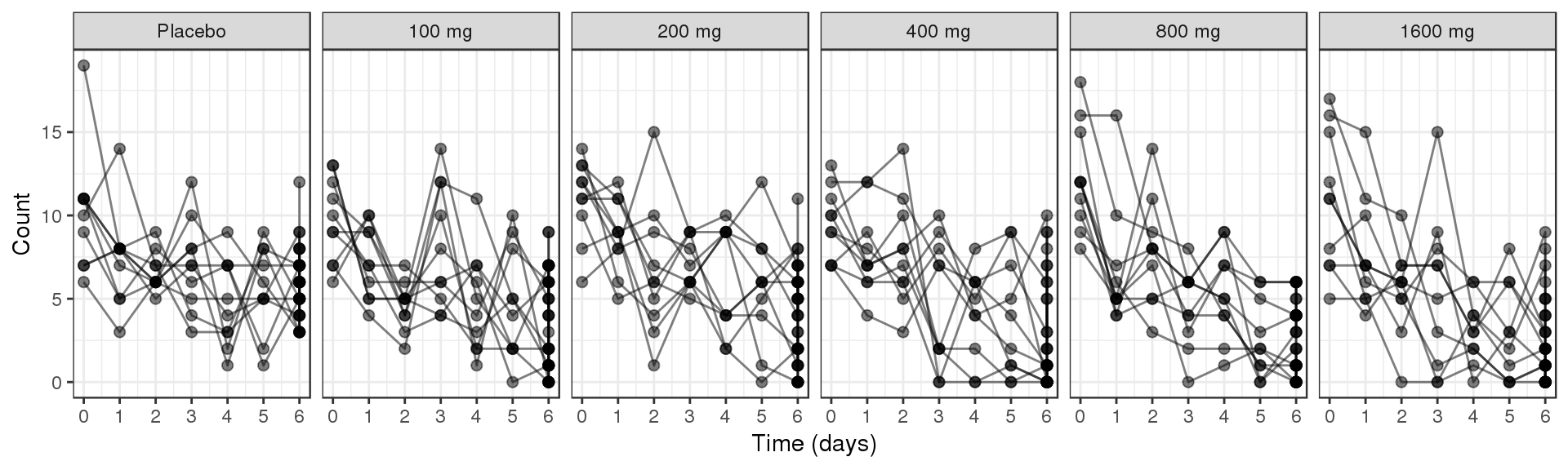
Explore irregularities in profiles
Plot individual profiles in order to inspect them for any irregularities. Inspect the profiles for outlying data points that may skew results or bias conclusions.
Count over time, faceted by individual, individual line plots
data_to_plot <- my.data[my.data$CMT==4,]
gg <- ggplot(data = data_to_plot,
aes(x=CYCLE,y=LIDV))+theme_bw()
gg <- gg + geom_point(size=2) + geom_line(aes(group = ID))
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Time (days)")+ scale_x_continuous(breaks = seq(-1,8,1))
gg <- gg + facet_wrap(~ID+TRTACT,ncol = length(unique(data_to_plot$ID))/length(unique(data_to_plot$DOSE)) )
gg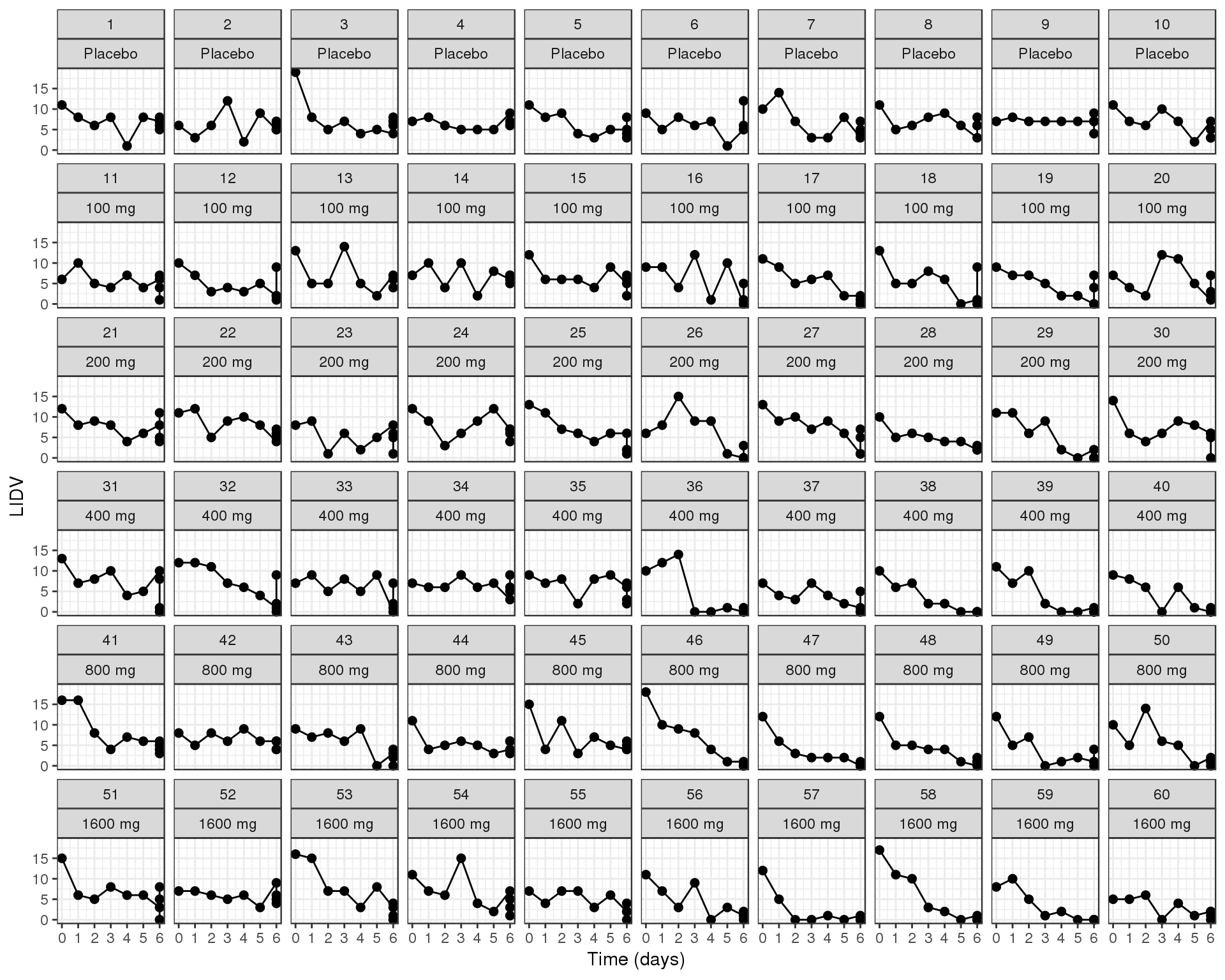
Explore covariate effects on PD
(coming soon)
Explore Dose-Response Relationship
Count vs Dose, colored by time, median, 5th & 95th percentiles by nominal time
data_to_plot <- my.data[my.data$CMT==4&my.data$DAY_label%in%c("Baseline","Day 1","Day 3","Day 5"),]
data_to_plot$TRTACT <- factor(data_to_plot$TRTACT, levels = rev(levels(data_to_plot$TRTACT)))
gg <- ggplot(data = data_to_plot,
aes(x=DOSE,y=LIDV, color = DAY_label, fill = DAY_label))+theme_bw()
gg <- gg + stat_summary(geom="errorbar",
fun.data=function(y){
data.frame(y=median(y),
ymin=quantile(y,0.025),
ymax=quantile(y,0.975))
}, size = 1, width = 0.2, position = position_dodge(50))
gg <- gg + stat_summary(geom="point", fun.y=median, size = 2, position = position_dodge(50))
# gg <- gg + geom_boxplot(aes(group = interaction(DOSE,DAY_label)),fill = NA)
# gg <- gg + stat_summary(geom="line",fun.y=median, size = 1)
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Dose (mg)") + scale_x_continuous(breaks = unique(data_to_plot$DOSE))
gg <- gg + ylab("Count")
gg <- gg + geom_smooth( method = "glm",method.args=list(family=poisson), position = position_dodge(50))
gg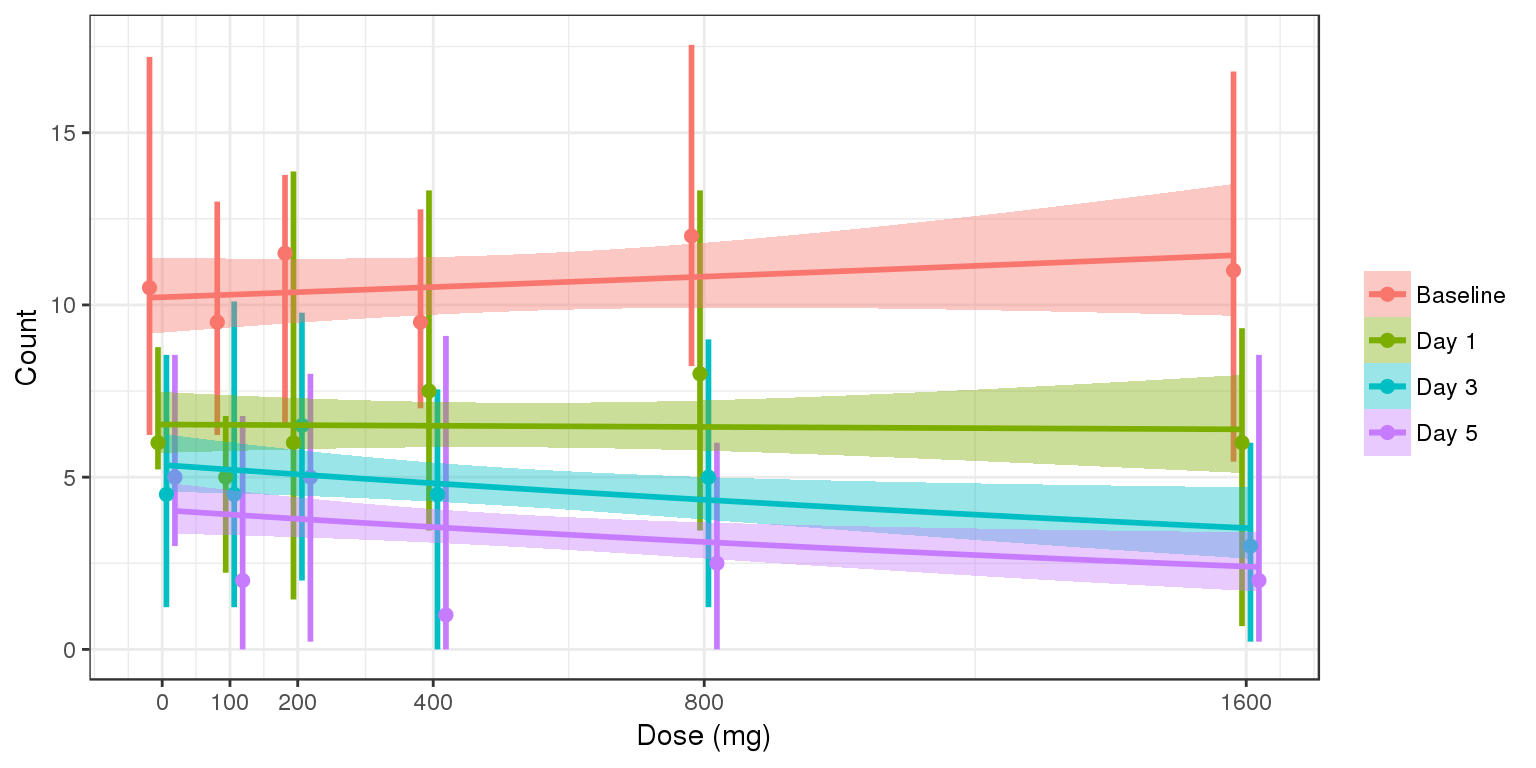
Count vs Dose, faceted by Time, boxplots by nominal time
data_to_plot <- my.data[my.data$CMT==4&my.data$DAY_label%in%c("Day 5"),]
gg <- ggplot(data = data_to_plot,
aes(x=DOSE,y=LIDV))+theme_bw()
gg <- gg + geom_boxplot(aes(group = DOSE))
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Dose (mg)")
gg <- gg + facet_grid(~DAY_label)
gg <- gg + ylab("Count")
gg <- gg + geom_smooth( method = "glm",method.args=list(family=poisson), color = "black")
gg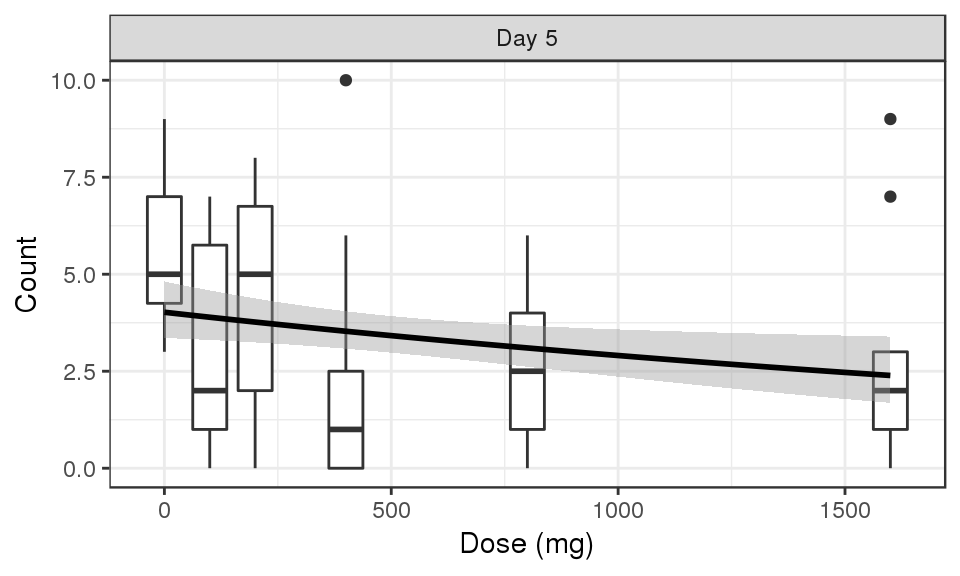
Count vs Dose, faceted by Time, boxplots by nominal time
data_to_plot <- my.data[my.data$CMT==4&my.data$DAY_label%in%c("Baseline","Day 1","Day 3","Day 5"),]
gg <- ggplot(data = data_to_plot,
aes(x=DOSE,y=LIDV))+theme_bw()
gg <- gg + geom_boxplot(aes(group = DOSE))
gg <- gg + guides(color=guide_legend(""),fill=guide_legend(""))
gg <- gg + xlab("Dose (mg)")
gg <- gg + facet_grid(~DAY_label)
gg <- gg + ylab("Count")
gg <- gg + geom_smooth( method = "glm",method.args=list(family=poisson), color = "black")
gg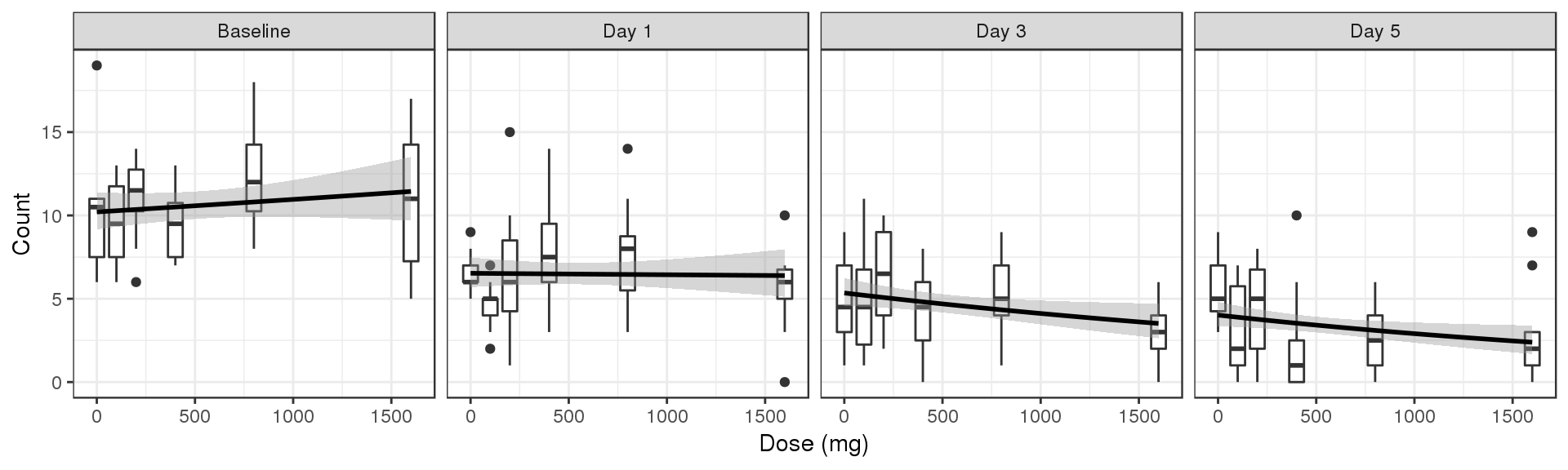
Percent of subjects by Count
data_to_plot <- my.data[my.data$CMT==4&my.data$DAY_label%in%c("Day 5"),]
# data_to_plot$DAY_label <- factor(data_to_plot$DAY_label, levels = rev(sort(unique(data_to_plot$DAY_label))))
# data_to_plot$TRTACT<- factor(data_to_plot$TRTACT, levels = rev(levels(data_to_plot$TRTACT)))
data_to_plot$LIDV <- factor(data_to_plot$LIDV, levels = rev(sort(unique(data_to_plot$LIDV))))
gg <- ggplot(data = data_to_plot, aes(x = factor(DOSE), fill = LIDV)) + theme_bw()
gg <- gg + geom_bar(position = "fill") + scale_y_continuous(labels = scales::percent)
gg <- gg + ylab("Percent of Subjects") + xlab("Dose (mg)") + guides(fill=guide_legend(""))
gg + facet_grid(.~DAY_label) + scale_fill_grey(start = 0.8, end = 0.2, breaks = rev(seq(0,18,6)))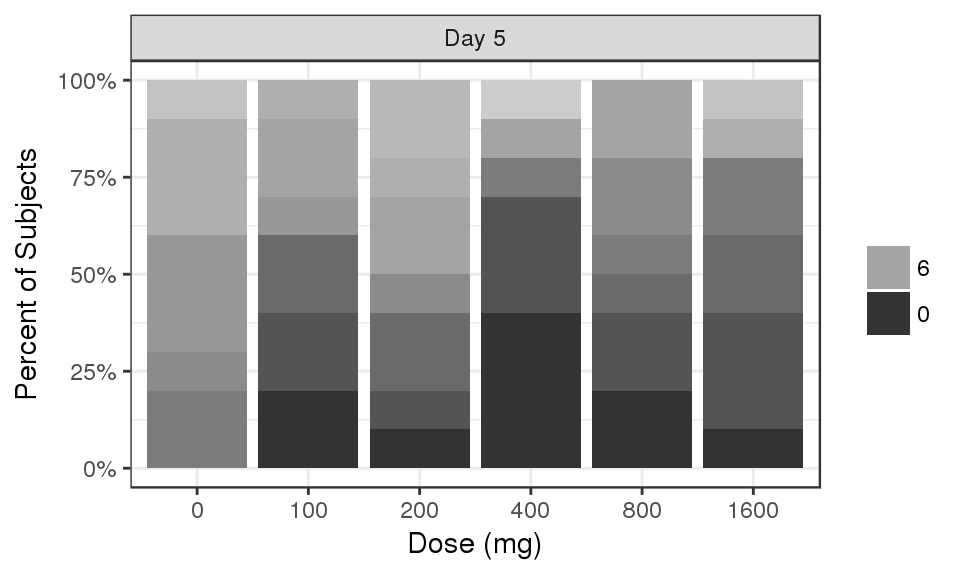
Percent of subjects by Count, faceted by time
data_to_plot <- my.data[my.data$CMT==4&my.data$DAY_label%in%c("Baseline","Day 1","Day 3","Day 5"),]
# data_to_plot$DAY_label <- factor(data_to_plot$DAY_label, levels = rev(sort(unique(data_to_plot$DAY_label))))
# data_to_plot$TRTACT<- factor(data_to_plot$TRTACT, levels = rev(levels(data_to_plot$TRTACT)))
data_to_plot$LIDV <- factor(data_to_plot$LIDV, levels = rev(sort(unique(data_to_plot$LIDV))))
gg <- ggplot(data = data_to_plot, aes(x = factor(DOSE), fill = LIDV)) + theme_bw()
gg <- gg + geom_bar(position = "fill") + scale_y_continuous(labels = scales::percent)
gg <- gg + ylab("Percent of Subjects") + xlab("Dose (mg)") + guides(fill=guide_legend(""))
gg + facet_grid(.~DAY_label) + scale_fill_grey(start = 0.8, end = 0.2, breaks = rev(seq(0,18,6)))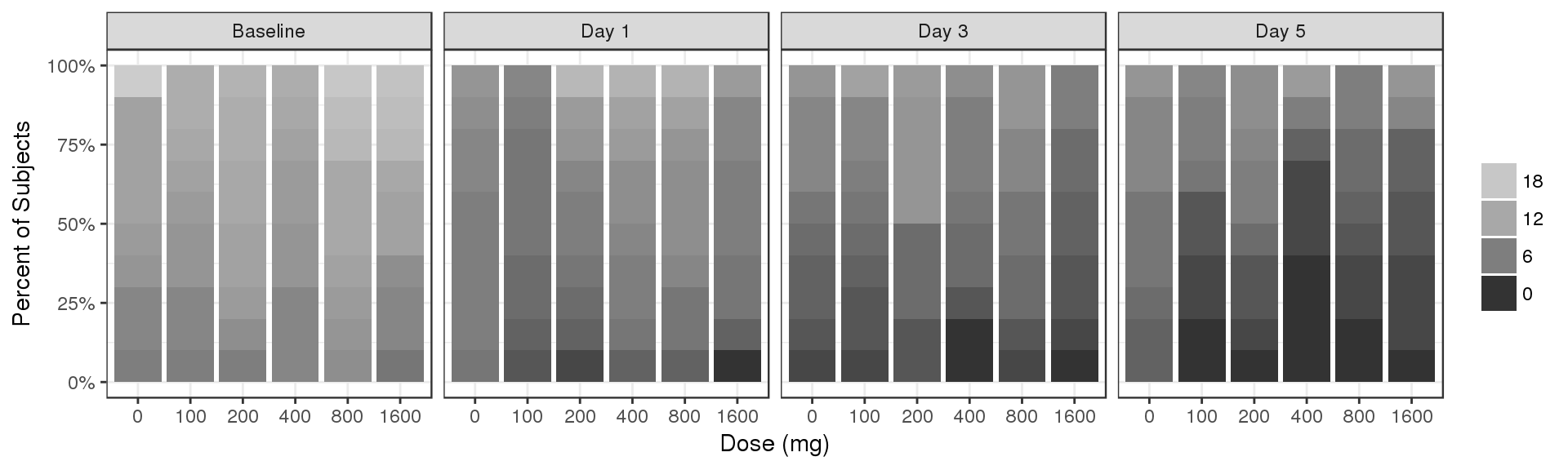
R Session Info
sessionInfo()## R version 3.4.3 (2017-11-30)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Red Hat Enterprise Linux Server 7.4 (Maipo)
##
## Matrix products: default
## BLAS/LAPACK: /CHBS/apps/intel/17.4.196/compilers_and_libraries_2017.4.196/linux/mkl/lib/intel64_lin/libmkl_gf_lp64.so
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] grid stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] DT_0.2 RxODE_0.6-1 bindrcpp_0.2 haven_1.1.0
## [5] readr_1.1.1 readxl_1.0.0 xtable_1.8-2 tidyr_0.7.2
## [9] caTools_1.17.1 zoo_1.8-0 dplyr_0.7.4 ggplot2_2.2.1
## [13] gridExtra_2.3
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.14 cellranger_1.1.0 compiler_3.4.3 pillar_1.0.1
## [5] plyr_1.8.4 bindr_0.1 forcats_0.2.0 bitops_1.0-6
## [9] tools_3.4.3 digest_0.6.13 jsonlite_1.5 memoise_1.1.0
## [13] evaluate_0.10.1 tibble_1.4.1 gtable_0.2.0 lattice_0.20-35
## [17] pkgconfig_2.0.1 rlang_0.1.6 rex_1.1.2 yaml_2.1.16
## [21] stringr_1.2.0 knitr_1.18 hms_0.4.0 htmlwidgets_0.9
## [25] rprojroot_1.3-1 glue_1.2.0 R6_2.2.2 binom_1.1-1
## [29] rmarkdown_1.8 reshape2_1.4.3 purrr_0.2.4 magrittr_1.5
## [33] codetools_0.2-15 backports_1.1.2 scales_0.5.0 htmltools_0.3.6
## [37] rsconnect_0.8.5 assertthat_0.2.0 colorspace_1.3-2 labeling_0.3
## [41] stringi_1.1.3 lazyeval_0.2.1 munsell_0.4.3 markdown_0.8